Joining two relationships with Scala and Spark
In this post we are going to analyze how we can read two relationships (R and S) from a csv file and then how we can compute the following relational operator in the most efficient way in Scala and Spark: R(a,b) ⋈ S(a,c). This is no more than a join operation on two tables. We are going to experiment with the built-in join methods of the Spark data structures (RDDs, DataSets and DataFrames) and we will also implement two manual ways: a simple one and one that groups and repartitions the records before the actual join. For each method we are going to count the miliseconds that the program needed to run and we will try to explain these results.
Note: Before we begin the source code displayed below can be found here.
Specifications
Let’s specify what we have to do here:
The given input data would be a text file and each of its lines have the following format:
Example:
R,2,a
R,4,b
R,3,a
R,5,b
R,6,a
R,7,b
S,2,Z
S,5,W
S,2,X
S,4,Y
R,2,g
S,7,w
The output of the program would be a message on the screen for each of the solutions implemented that will contain the time (in ms.) that was needed for this solution to run and the number of results.
Finally, the joined relation should be written in different output folders in the disk.
[1] The RDD way
The first way to go is using the RDD structure of the Spark framework. In this way, we need to read the input file using the SparkContext into an RDD. Then, we have to split this RDD into two new RDDs that each of them will only contain records from one relation (either “R” or “S”) and then we will use the built-in method join() that RDDs have. Following this logic, I reached to the following solution:
def joinUsingRDDs(sc: SparkContext, inputFile: String, outputDir: String): Unit = {
val inputRDD = sc.textFile(inputFile, 2)
val s_relation = inputRDD.filter(line => line.charAt(0) == 'S')
.map(line => line.split(","))
.map(line => (line(1), line(2)))
val r_relation = inputRDD.filter(line => line.charAt(0) == 'R')
.map(line => line.split(","))
.map(line => (line(1), line(2)))
val startTime = DateTime.now(DateTimeZone.UTC).getMillis()
val joined = r_relationship.join(s_relationship)
println("[RDD join] \t Time: " + (DateTime.now(DateTimeZone.UTC).getMillis()-startTime) + "miliseconds \t Number of results: " + joined.count())
joined.saveAsTextFile(outputDir)
}[2] The DataFrame way
The second way to go is using the DataFrame structure of the Spark framework. In this way, we need to read the input file using the SparkSession into a DataFrame. Then, we have to split this DataFrame into two new DataFrames that each of them will only contain records from one relation (either “R” or “S”) and then we will use the built-in method join() that DataFrames have. As you can understand the logic is very similar to the one of the RDD-solution. Bellow, is the solution using DataFrames:
def joinUsingDFs(spark: SparkSession, inputFile: String, outputDir: String): Unit = {
val df = spark.sqlContext.read.option("header", "false").option("delimiter", ",").csv(inputFile)
val cols = Seq[String]("_c1", "_c2")
val s_df = df.filter("_c0 == 'S'")
.select(cols.head, cols.tail: _*)
.toDF(Seq("key", "valueS"): _*)
val r_df = df.filter("_c0 == 'R'")
.select(cols.head, cols.tail: _*)
.toDF(Seq("key", "valueR"): _*)
val startTime = DateTime.now(DateTimeZone.UTC).getMillis()
val joined = r_df.join(s_df, Seq("key"))
println("[DataFrame join] \t Time: " + (DateTime.now(DateTimeZone.UTC).getMillis()-startTime)
+ " miliseconds \t Number of results: " + joined.count())
joined.write.csv(outputDir)
}[3] The DataFrame with SQL way
The third way is using the DataFrame structure of the Spark framework, but this time along with the SQL method of SparkSession. Again, at first we need to read the input file using the SparkSession into a DataFrame. Then, we have to split this DataFrame into two new DataFrames that each of them will only contain records from one relation (either “R” or “S”) and then we will use the SQL method provided by Spark to enter our SQL query which will join the two relations.
Note: We could have used SQL to also split the DataFrame, but since we start to count time just before we join the two relations, it would have made no difference
def joinUsingSQL(spark: SparkSession, inputFile: String, outputDir: String): Unit = {
val df = spark.sqlContext.read.option("header", "false").option("delimiter", ",").csv(inputFile)
val cols = Seq[String]("_c1", "_c2")
df.filter("_c0 == 'S'")
.select(cols.head, cols.tail: _*)
.toDF(Seq("key", "valueS"): _*)
.createOrReplaceTempView("s_table")
df.filter("_c0 == 'R'")
.select(cols.head, cols.tail: _*)
.toDF(Seq("key", "valueR"): _*)
.createOrReplaceTempView("r_table")
val startTime = DateTime.now(DateTimeZone.UTC).getMillis()
val joined = spark.sql("SELECT s_table.key, s_table.valueS, r_table.valueR FROM s_table INNER JOIN r_table ON s_table.key = r_table.key”)
println("[DataFrame join] \t Time: " + (DateTime.now(DateTimeZone.UTC).getMillis()-startTime)
+ " miliseconds \t Number of results: " + joined.count())
joined.write.csv(outputDir)
}[4] The DataSet way
The fourth way and the last built-in way we are going to explore is using the DataSet structure of the Spark framework. At first we need to read the input file using the SparkSession into a DataSet. Then, we have to split this DataSet into two new DataSets that each of them will only contain records from one relation (either “R” or “S”) and then we will use the joinWith() method of DataSet to join the two sets.
The logic is again similar to what we have already seen. The only difference is the data structures that are used in each case.
def joinUsingDS(spark: SparkSession, inputFile: String, outputDir: String): Unit = {
val inputData = spark.read.option("header", "false").option("delimiter", ",").textFile(inputFile)
val s_relation = inputData.filter(line => line.charAt(0) == 'S')
.map(line => line.split(","))
.map(line => (line(1), line(2)))
val r_relation = inputData.filter(line => line.charAt(0) == 'R')
.map(line => line.split(","))
.map(line => (line(1), line(2)))
val startTime = DateTime.now(DateTimeZone.UTC).getMillis()
val joined = s_relation.joinWith(r_relation, s_relation("_1") === r_relation("_1"))
println("[DataFrame join] \t Time: " + (DateTime.now(DateTimeZone.UTC).getMillis()-startTime)
+ " miliseconds \t Number of results: " + joined.count())
joined.write.json(outputDir)
}[5] The manual way
The this way we will read the data into a RDD structure and then without using any built-in method we will group the records by their keys, then for each key we will take all the values, filter them by their relation symbol column and combine each record of the one relation with all the records of the other.
This is the simplest manual way to perform a join and it would be interesting to see how efficient this way is compared to the other built-in ways.
def joinManually(sc: SparkContext, inputFile: String, outputDir: String): Unit = {
val myData = sc.textFile(inputFile,2)
val startTime = DateTime.now(DateTimeZone.UTC).getMillis()
val joined = myData.map(line => line.split(","))
.map(line => (line(1), (line(0), line(1), line(2))))
.groupByKey()
.flatMapValues(tuples => tuples.filter(tuple => tuple._1 == "R")
.flatMap(tupleR => tuples.filter(tuple => tuple._1 == "S")
.map(tupleS => (tuple._3,tuple3._3))))
println("[DataFrame join] \t Time: " + (DateTime.now(DateTimeZone.UTC).getMillis()-startTime)
+ " miliseconds \t Number of results: " + joined.count())
joined.saveAsTextFile(outputDir)
}[6] The manual way #2
There is another way to handle the join task using a different optimised technique. We are going to use the value-to-key conversion design pattern. That means that in the mapper phase, instead of simply emitting the join key as the intermediate key, we instead create a composite key consisting of the join key and the tuple id (from either R or S). At this point, we must define the sort order of the keys to first sort by the join key, and then sort all tuple ids from R before all tuple ids from S - and we are lucky because this is what Spark does by default. The tricky part is that we must define a custom partitioner to pay attention to only the join key, so that all composite keys with the same join key arrive at the same reducer.
Following this procedure results in having all the tuples from S with the same join key to be encountered first, which the reducer can keep in memory. Then, as the reducer processes each tuple from S, it is crossed with all the tuples from R, which will finally give us the desired result.
Of course, we are assuming that the tuples from R (with the same join key) will fit into memory, which is a limitation of this algorithm.
def joinManually(sc: SparkContext, inputFile: String, outputDir: String): Unit = {
val myData = sc.textFile(inputFile,2)
val startTime = DateTime.now(DateTimeZone.UTC).getMillis()
val joined = myData.map(line => line.split(","))
.map(line => ((line(1), line(0)), (line(0), line(2))))
.repartitionAndSortWithinPartitions(new CustomPartitioner(2))
.map(line => (line._1._1, line._2))
.groupByKey()
.flatMap(tuple => {
var listOfR : List[String] = List()
tuple._2.map(record => {
if (record._1 == "R")
listOfR = listOfR :+ record._2
else
listOfR.map(valueR => (tuple._1, (valueR, record._2)))
})
})
.filter(_.isInstanceOf[List[(String, (String, String))]])
.map(_.asInstanceOf[List[(String, (String, String))]])
.flatMap(x => x)
println("[DataFrame join] \t Time: " + (DateTime.now(DateTimeZone.UTC).getMillis()-startTime) + " miliseconds \t Number of results: " + joined.count())
joined.saveAsTextFile(outputDir)
}
class CustomPartitioner(numParts: Int) extends Partitioner {
override def numPartitions: Int = numParts
override def getPartition(key: Any): Int = {
key match {
case tuple @ (a: String, b: String) => Integer.valueOf(a) % 2
}
}
}Conclusion
Running all the implementations described above, for the given input text file will produces this output:
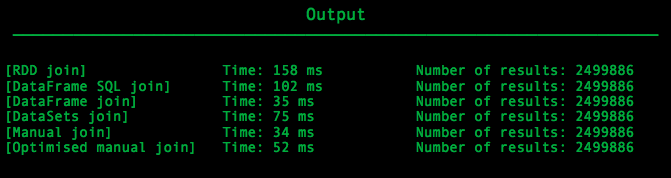
As we can see the slowest method was the RDD structure which the expected result. RDDs is an old structure of Spark framework, a stable one of course, but on the other hand there are newer structures like DataFrames and DataSets that use better optimisations under the hood.
The second slowest method was the usage of SQL to join the two tables. This method was not so slow and it had small differences with the best solution. Maybe this is a good indication that using SQL on DataFrames may not be the most optimal way to perform actions on data, but still Spark gives its users a decent way to use the well-known SQL queries over one of his best structures.
DataFrame and DataSet data structures are the newer structures of Spark that use a lot of optimisations to achieve the best possible performance. As we can see on our output, in our example DataFrames performed better than the DataSets but this may not be always true.
Surprisingly, our simple manual way to join the two tables was the best solution. Maybe just filtering an RDD to split it into two and then directly cross each record of the first one with all the records of the second one is an optimal solution. Maybe, this logic become more slow as the data become bigger and does not fit in the main memory. At this scenario, the optimisations of Spark built-in data structures may be more efficient.
Last but not least we have the optimised solution that works as we described above. This solution performed better than the DataSets and was just a little slower than the join with the DataFrame structure and the simple manual join. This slowness may have occurred due to the necessary transformations that transformed our joined result in the desired form or due to my inexperience in Scala programming. Still, this solution is one of the most optimal ones under the limitation that all the records of each key of the smallest relation will fit into main memory, which was our case.
